Digits: Two Reports on New Units of Scholarly Publication
Skip other details (including permanent urls, DOI, citation information): This work is licensed under a Creative Commons Attribution 4.0 International License. Please contact [email protected] to use this work in a way not covered by the license.
For more information, read Michigan Publishing's access and usage policy.
A New Unit of Publication
The potential of software containers for digital scholarship
Picture a web-based digital publication. Whether it contains an interactive map, a network visualization, a curated collection of born-digital objects, or other multi-modal expression, chances are this project is built upon layers of technological systems or “stacks.” A stack is an assemblage of software that forms the operational infrastructure behind the project itself. Industry professionals often speak of the LAMP stack, the MEAN stack, and Ruby on Rails and, indeed, these technologies are the cornerstone of the web.[2]
A stack's seeming ubiquity, however, creates an illusion of monolithic, laborless setup and configuration underpinning the data modeling and public facing layers of production that most digital humanities scholars and web developers tend to focus on. Anyone who has attempted to maintain, preserve, or replicate a digital project knows, however, that the deepest layers of any server stack can have a profound impact on how algorithms run and how information displays. A given operating system will immediately enable or prohibit certain software; one’s choice of database can erase an important difference between the two types of data; failing to apply a software patch behind schedule can, in the words of Deltron, "crash your whole computer system and revert you to papyrus."[3]
These challenges have given rise to two widespread paradigms of support: dedicated servers with full time, in-house system administrators or large-scale, cloud-based vendors who offer varying levels of stack flexibility and system administration support. In a higher education IT or on cheap commodity cloud providers, one is likely to find flexibility (say, a virtual machine with no predetermined operating system or software) with little or no support, or a well supported software stack designed for a narrow set of use-cases. Within this context, playing around with ideas, creating experimental prototypes, reproducing another’s work to interrogate it—or collectively, “sandboxing”—is particularly difficult.
In the face of these challenges, an approach called software containerization is becoming increasingly popular. Containerization offers a potential solution to the primary challenges of maintenance, reproducibility, and preservation for web-based digital scholarship, but also necessitates a significant departure from the current status quo. In December 2016, our working group received funding from the Andrew W. Mellon Foundation for “Digits: a Platform to Facilitate the Production of Digital Scholarship,” an 18-month project to survey the use of container technologies in scholarly publication, assess the needs of researchers producing web-centric scholarship, and develop blueprints for a platform to facilitate those needs. The first stage of our investigations has been to author an environmental scan of software containers in scholarly contexts with two focal research questions:
- How are containers being discussed and adopted in the academic research contexts?[4]
- Which aspects of containerization have not yet been fully explored in the context of digital scholarship?
In Section 1: Technical Background, we begin by providing a short introduction to container technology. This opening section attempts to introduce container-based approaches in a manner accessible to an imagined reader without deep technical or server “back-end” experience. Section 2: Containers in Academia provides a review of formal and informal conversations about software containers across a variety of disciplines. As a substantive body of published scholarship demonstrates, containerization has seen more use in high-performance computing (HPC) and the other scientific contexts than it has in digital humanities. Section 3: Full-Stack Scholarship focuses on the prevalent but as-yet-unrealized idea that containers might come to function as standalone publications (self-contained, as it were), with each software container encapsulating a new unit of publishable work comparable to an article or a monograph.
This document focuses on the use and potential for containers within the academic research community, however the development of software containers (with some exceptions discussed below) is primarily driven by industrial needs and resources. We cannot hope to cover all of that activity here, as it is already challenging to track everything happening with containers in academia.[5]
Technical Background
This section provides a semi-technical introduction to software containers, intended to provide a high-level overview of important and relevant technical concepts of software containers without getting too bogged down in the system-level details of any particular implementation. The section includes a comparison between containers and virtual machines, a discussion of five relevant container technologies, and finally a brief introduction to orchestration. The technical background also includes a short consideration of the social and organizational significance of containers, drawing from their impact in industry and the implications for research workflows and infrastructure.
What are Containers?
Software containers are a suite of technology components for Linux-based operating systems that enable the isolation of computational processes. More specifically, containers are an instance of operating-system-level virtualization.[6] Many of the technologies for isolating resources that make containers possible (LVM, chroot, cgroups) have existed for many years, but have only recently been integrated into easy to use interfaces, the most popular being Docker.[7] Docker is by far the most widely adopted container technology, but there are alternatives such as rkt[8] or singularity (Kurtzer et al. 2017).
The recent popularity of containers, namely Docker, stems from the capability to “package software into standardized units for development, shipment and deployment.”[9] This statement from Docker’s marketing materials can be broken down to illustrate why containers have seen a dramatic rise in popularity and public discussion since June 2014 when Docker 1.0 was released.
- Package software: Containers make it easy to bundle or envelop software applications and services along with their software dependencies to create an executable thing with clearly defined boundaries.
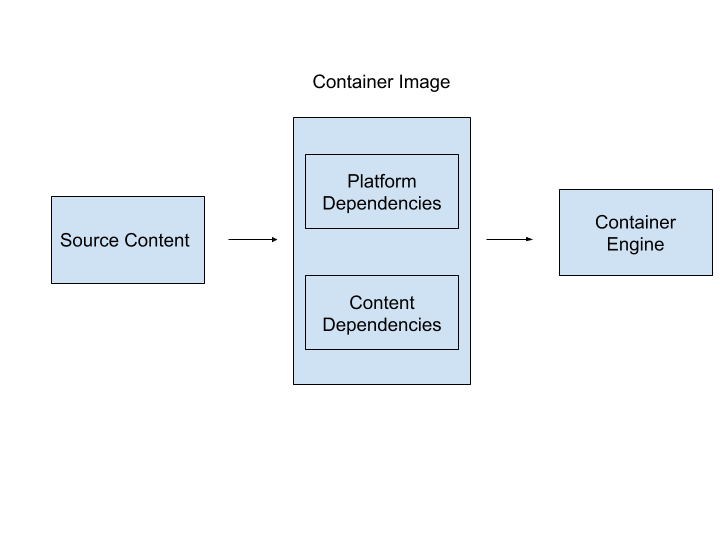
- Standardized unit: The thing-ness of software containers is embodied in the container image, a file format for bundling the components of a software application including the code, compiled binaries, configuration files, data files, dependencies, or anything else needed for the execution of the contained software application. This standardization make possibly the fast and easy movement of applications across platforms.
- Development, shipment and deployment: Containers have standardized interfaces for execution, which means that software contained within them will execute consistently on any system capable of running containers (assuming the same underlying container technology). This standardization enables the contained software to run in many different environments without dealing with significant overhead related to software dependencies and system configuration. Packaging and standardization allows containers to be portable across environments with each environment having far fewer software dependencies, namely the container runtime. Importantly, the environment used to develop the application (perhaps on a developer’s laptop) can be identical to the production environment (perhaps on a commodity cloud provider like Amazon Web Services). The bundling of containers into standardized units makes moving the contained application from the developer’s laptop to the cloud deployment (“shipping”) easier and less likely to introduce bugs because the runtime environment is (nearly) identical.
In order to fully understand software containers, one must widen the scope of one’s idea of “software.” For many users, software applications might bring to mind software such as Microsoft Word or apps for smartphones. While it is theoretically possible to put client side applications with graphical user interfaces into a container, the technology was not necessarily designed for such use cases;[10] for purposes of this paper it is most reasonable to think of a container as a mechanism for packaging a server. Nearly all of the software applications being put into containers run on the Linux operating system and are back-end applications like web-servers, databases, business application servers, middleware, etc.
Containers’ immediate appeal in server administration and web development is due to three central features (or affordances): performance, encapsulation, and portability. Containers have fast performance because they are system processes, not separate operating systems, which leads to very fast “boot up” times and low resource (memory, CPU) overhead. Such speed is possible because containers share many of the operating system resources with the host computer. Empirical analysis has shown (Felter et al. 2015; Hale et al. 2016; Le and Paz 2017) that for CPU and memory tasks, containers performed nearly as fast as native hardware and faster than virtual machines (discussed more below).
Encapsulation is the idea of circumscribing all the code an application needs to execute in a single logical unit. All too often, the process of server setup is one of saying “I need to install application A, but to install that I need to install library B, but to install B I need to install library C.” The resulting state, “dependency hell,” results in profound frustration. Detailed installation instructions can help for identical environments but may leave out assumed information on taken-for-granted dependencies. Conditions like these are widespread in scholarly computing, where reproduction of others’ code/results is considered especially important. When applications are developed inside a container, the only external dependency becomes the container runtime environment itself.
Portability builds on top of encapsulation, as containers are portable across different host environments. The container engine performs as an infrastructural gateway, not unlike an actual shipping container, capable of executing a wide variety of software conforming to a standard interface (Egyedi 2001). Portability is made possible through a container image. A container image is an immutable file format within which a contained application’s dependencies are bundled together. Container images can be executed creating a “live” or running instance of a particular image. As a result, one can easily have two containers running from the same image, reducing configuration overhead. For example, a developer might create an image that contains a web server and a default folder for the website that server will host. Multiple websites with the same dependencies could be hosted by launching two or three or even more containers from that image. This ease of reuse is the key to a container’s portability. Locally, an image can operate as a base for many containers.
Containerization is not without detractors and skeptics. Foremost, the sheer exuberance for containers makes some wonder how much of the attention is mere hype. Academic IT tends to be especially wary of committing to a technology that could become obscure within a 3-to-5 year timescale. Further, containers are only supported on Linux, both for the host and the system running inside the container. In contexts where Linux is already the preferred operating system (e.g., server administration and web development), containerization offers more immediate gains than losses. The small performance hit has been seen as a worthwhile price to pay for the benefits of encapsulation and portability for all except the most extreme computationally intensive tasks.
Containers vs. Virtual Machines
How are containers different from virtual machines? Virtual machines envelope more layers of the computing environment, going deeper down into the system and emulating computer hardware. Virtual machines run operating systems where containers run software applications. .
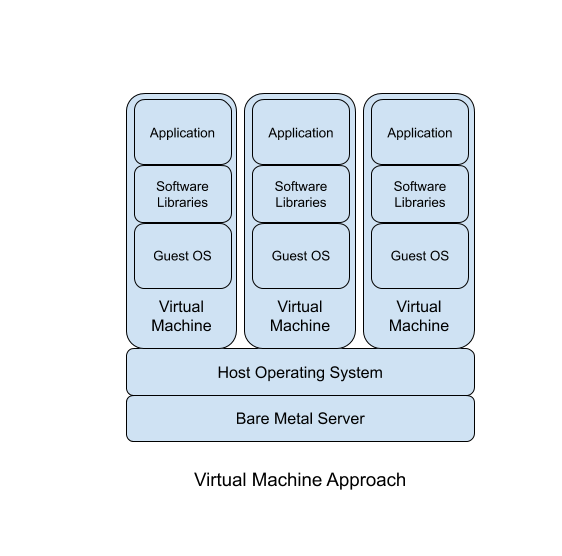
The focus on applications means containers are much lighter weight than virtual machines. Containers do not need to “boot up” because they draw more resources from the host operating system and share resources with other containers running on the system; they are already “booted up”. This efficiency means the overhead of containers is much smaller than virtual machines. The performance and efficiency of containers (vs. virtual machines) comes at a reduced flexibility for the kinds of software applications that can be run inside of containers. Where a virtual machine can theoretically run any operating system capable of running on the virtualized hardware, containers can only run software developed to run on Linux.
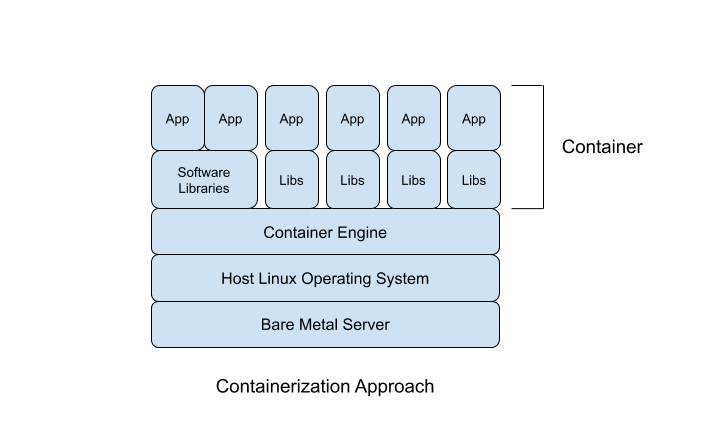
Containers are not a replacement for virtual machines; they function at a different level of abstraction. In many deployments, containers are being used inside virtual machines (especially in the commodity cloud where everything is virtualized) as a means of using computational resources as efficiently as possible. For example, if an enterprise needs four redundant web servers, rather than having four heavy virtual machines each only running a web server, they can have a single virtual machine (or two for an extra layer of redundancy) running multiple containers for each web server. This means running (and paying for) two virtual machines instead of four. The efficient use of computational resources is one of the value propositions of software containers, especially for enterprise applications like web-servers that are not very computationally intensive.
A seasoned system administrator might say “I don’t need containers to run multiple web servers, I can just run them myself!” and they would be correct. However, the portability and standardization of containers, not to mention the software ecosystem that has emerged to control the creation, deployment, and management of containers, makes running applications and services several orders of magnitude easier than it has ever been in the past.[11]
Container Technologies
Technology | Launch date | License | Comments |
(“Docker” Enterprise 3.0) (runc)[12] | June 2014 | Apache 2.0 | Most popular container technology, runs on 15% of hosts monitored, according to one study last updated in April 2017. The annual Portworx Container Adoption Survey suggests that Docker’s reputation has steadily declined since 2017.[13] |
(“CoreOS rkt”)[14] | July 2016 | Apache 2.0 | Under active development. Appears to be growing in popularity, especially given the fact it works with Kubernetes and is not Docker. |
April 2016 | 3 clause BSD | A container technology developed specifically for scientific computing. Has same advantages of industry containers (portability, reproducibility, environmental encapsulation) with additional security and an execution model better suited to HPC environments. | |
December 2015 | 3 clause BSD | Another container technology specifically for HPC. Pre-cursor to Singularity. Development may have slowed or stalled. | |
June 2015 | n/a | An industry led standardization effort. Current focus is a Runtime Specification and an Image Specification for executing and bundling containers. Designed around a runtime and format donated by Docker (runc).[18] |
This list of container technologies in the table above is not exhaustive[19] but represents the most popular or most relevant technologies for this discussion (as of 2017). As of this writing, Docker is the de facto standard for the industrial application of containers. It should be noted that Docker is not simply a container technology, but rather a suite of technologies including a container runtimev(runc), specification formats (Dockerfile, docker-compose.yml), image server (Docker Hub), and orchestration system (Swarm).[20]
The academic community has developed its own alternatives to industrial software containers. Shifter and, more popular, Singularity. These technologies have emerged from the unique needs of high performance computing (HPC) where the computational workloads aren’t long-running, low overhead web services, but rather computationally intensive (but bounded) data processing or simulation jobs.[21] In high performance computing, containers are less of a solution to optimal utilization of resources (they are already good at this) and more focused on leveraging containers to manage the complexities of scientific software. Furthermore, the security model of Docker, running with elevated user privileges, is fundamentally incompatible with the current security model of HPC system where users typically have very few privileges to install software or configure the system.
The advantage of containers for HPC is the ability to support user defined images (Douglas M. Jacobsen and Canon 2015; D. M. Jacobsen and Canon 2016): where the work and responsibility of managing the software and its dependencies is pushed on to the researcher or scientist to configures their software environment as they please. Once they have added all of the needed content (dependencies, data, code, etc.) they can move the container image to a shared resource, most often a high performance research computing cluster, and run their computation without the need for the system administrators of the shared resource to install and configure the researcher’s software. The appeal of operating containers at a non-admin privilege level is substantial, and the demand for secure containerization should not be underestimated.
The vision for academic computing proposed by Singularity containers leverages the affordances of container technology (encapsulation and portability) while also integrating into existing technical infrastructure and workflows of research computing.
Additionally, Singularity enables integration with existing job scheduling systems used by the research computing community like the Slurm Workload Manager.[22] Singularity containers in essence become self-contained binary applications with a single dependency, the container runtime, instead of the traditional HPC use case of managing the gordian knot of interdependencies for all the scientific software needed by the many users of a shared computing facility.
Orchestration
While containers in and of themselves have reconfigured the landscape of system administration, the most significant impact has been the coupling of software containers with new breeds of orchestration systems. Orchestration, broadly, is the automated management of systems and services within some kind of computational ecosystem.[23] While orchestration is not a new concept, container orchestration has enabled the technology industry to provide services at a scale never before possible. Google’s Gmail web service (and many other Google services) are composed and managed in containers with orchestration (Verma et al. 2015). Orchestration systems like Kubernetes,[24] provide a set of abstractions to allow administrators to define the applications and services they want (and their redundancy) and then handles all the messy work maintaining that environment automatically.
The technological landscape of orchestration is rapidly changing, so any technical description given today would be immediately out-of-date. It is more important to know how orchestration changes the nature of the work of developing, deploying, and maintaining systems and services. Consider the difference between a chef cooking individual meals at a restaurant and the standardized meals prepared at a fast food restaurant. While the former focuses attention to each plate, the latter scales to “billions and billions served.” Container orchestration shifts the attention of the system administrator away from the deployment and management of individual systems towards collections of applications and services. With orchestration, the emphasis is less upon setting up a bare metal server or virtual machine with hands-on configuration, because this approach does not scale to hundreds or thousands or tens of thousands of instances. Instead, given a cluster of identical, minimally configured nodes (either bare metal or virtual machines), the system administrator leverages orchestration software and pre-configured containers to articulate an ecosystem at the scale of the data-center instead of the individual system. Much like fast food, this paradigm of service emphasizes certain kinds of use cases where bulk processing is more important than individuated attention.[25]
Orchestration forces an infrastructure level perspective and enables the management and operation of heterogeneous applications and services at scale. Installing and configuring software occurs once in the creation of a container image, which can then be replicated across a computing cluster. Orchestration for enterprise or industrial applications and service is related to the job scheduling in HPC discussed above, but the technologies, capabilities, and the kinds of workloads are very different (web servers and databases vs. computational modeling). Commodity cloud providers like Azure, Amazon, and Google offer containers as a service,[26] which is possible because of orchestration systems like Kubernetes (which is also offered as a service[27]).
Containers and orchestration posit radically new ways of managing computation. As such, new best practices, like The Twelve Factor App[28] are challenging traditional models for the architecture, development, and deployment of enterprise applications and services. These new paradigms, like microservices[29] and serverless computing,[30] are radical departures in the architecture, development, and deployment of enterprise applications and services. For example, Microsoft’s Azure Container Instances abstract away the infrastructural boilerplate of networking, filesystems, virtual machines, and server configuration, rending all of that work invisible to the user (invisible, but not eliminated). These systems are designed to execute (and bill) containerized applications on the order of seconds instead of days or weeks.[31] Containers move and consolidate specific forms of technical work, which has benefits in terms of labor efficiency, but may have broader implications as well.
The Significance of Containers
Docker’s logo is a whale carrying shipping containers. The developers and advocates of software containers liken their impact to that of shipping containers. This analogy is often used as a justification to skeptical systems or business administrators who are, rightly-so, risk averse when it comes to new technology. Just as shipping containers reduced the cost and simplified the logistics of moving material goods across the Earth (Levinson 2008), software containers made it easier to “ship” applications and services. When the means of executing software is standardized, infrastructure can be designed around a standard interface rather than a multitude of uniquely designed applications.[32]
Alongside these new technologies emerge new ways of working, sometimes called the “DevOps” philosophy (Clark et al. 2014). The term DevOps originated with efforts to break down the traditional siloes of development and operations. Instead engineers work “across the entire application lifecycle” and cultivate “a range of skills not limited to a single function.”[33] DevOps takes the programming, scripting, and automation abilities of developers and focuses those efforts on the operation of information infrastructure. Essentially the DevOps approach is one that automates much of the system administration performed by hand. More tritely, DevOps is about tools-for-managing-tools, which has resulted in a cambrian explosion of new tools and techniques for managing existing suites of tools for managing services like web servers, databases, or application servers.
Containers introduce not only a new set of tools to learn, but a whole new set of concepts and a philosophy of system administration. This conceptual change is perhaps the most significant, and disruptive, aspect of software containers. The affordances of portability and encapsulation also change how particular forms of IT work are done, and by whom. The analogy of the shipping container raises significant, and problematic, questions about labor and the visibility of work. These are questions we must keep at the forefront of the conversations around software containers for digital scholarship in the digital humanities.
Container use in Academia
Three topics emerged from our review of the published literature and ongoing conversations about containers in academic research contexts. First, we cover containers for software dependency management. A growing body of scholarship suggests that this is the most immediate and salient value proposition associated with containers for scholarly endeavors. Second, we cover containers for reproducible research. As with software dependency management, reproducibility seems to be a promising benefit of moving to a containerization approach. Third, we address the existing literature on using software containers for preservation. There is perhaps less consensus pertaining to the pros and cons of using containers in this way, but digital preservation is a crucial concern among many scholarly communities, so an assessment of how scholars have discussed this subject is warranted. Overall, this section describes how these mostly separate uses of containers lay the groundwork for beginning to imagine software containers as a new unit of publication.
Containers for Software Dependency Management
Much of the software dependency management conversation emerged from the HPC community, which has been dealing with the “dependency hell”[34] problem for decades. Dependency hell describes the problem of managing the menagerie of shared software packages or libraries that a particular application, especially a scientific application, requires in order to function. Software dependencies can be shared by multiple applications, which quickly creates a tangled morass of inter-dependencies. Dependency hell adds costs, both in terms of time and money, to the development, deployment, and redeployment of software; an especially potent problem for the management of scientific software.
The high performance computing community suffers, perhaps more than any other academic group, from challenges with software dependency management because research computing groups manage computational clusters as a service for researchers with varying computational needs and expertise. Because of this variability, many research computing clusters are centrally controlled, where system administrators, not the individual users, must manage the software configuration of the cluster. This workflow places system administrators squarely in the path of getting science done, which can create tensions when scientists need bespoke or highly customized software for their specific research. System administrators do not scale and complex software dependencies coupled with poorly designed scientific software results in dependency hell.
Software containers alleviate the challenges of dependency management, especially for the management of software in high-performance computing environments, by giving users the “privilege” of the administrative labor of installing software (Belmann et al. 2015; Moreews et al. 2015; Szitenberg et al. 2015; Chung et al. 2016; Devisetty et al. 2016; Hosny et al. 2016; Hung et al. 2016). Software containers, especially implementations like Singularity, allow for researchers to work with the software environment they like, as opposed to conforming to the standardized and secure software environment of the HPC cluster. Through portability and encapsulation, software containers afford a more robust environment for running scientific software without the burden of an exponentially growing list of software to support or secure. In theory, the only software dependency is the container runtime itself.
The portability and performance of containers allows a researcher to move quickly and easily from their laptop or desktop as soon as their research needs have outgrown their current resources.
Images can provide a complete, stable, and consistent environment that can be easily distributed to end users, thereby avoiding the difficulties that end-users commonly face with deep dependency trees. Containers have the further advantage of largely abstracting away the host system, making it possible to deliver a common and consistent environment on many different platforms, be it laptop, workstation, cloud instance or supercomputer. (Hale et al. 2016, 14).
Examples of this approach include projects like Bioboxes (Belmann et al. 2015), which is an effort to address the difficulties installing and maintaining software in bioinformatics using Docker containers with standardized interfaces. Bioinformatics relies upon complex and custom software creating usability problems that can inhibit the progress of science. Other efforts in the biosciences, such as the University of Pittsburgh’s Center for Causal Discovery (CCD) use preconfigured Docker containers to relieve difficulty configuring their causal modeling applications.[35] Instead of battling with the complexities of the Python to Java software bridge, researchers can just run their ready-to-run container (personal communication). The CCD container builds on top of the Jupyter Docker Stacks,[36] which are a collection of generic Docker containers with Jupyter Notebooks and other Python libraries for data science and scientific computing. The DHbox project[37] is using Docker containers to manage the complexity of installing popular digital humanities tools and create a ready-to-run “laboratory in the cloud” for research and teaching.[38]
Software containers for science are often framed as creating portable research environments or workbenches (Willis et al. 2017). The idea here is that all of the software dependencies necessary to do science are bundled up and made available to the researcher, who connect the data needed for doing their science. This offloads the headache of each researcher compiling and configuring software on their environment by allowing them to build on top of other (perhaps more experienced) effort to set up scientific software. In this sense, software containers allow for standardized workbench/lab-bench like computing environments for the scientists to do their work. This begs the question, if we can bundle scientific software environment can we bundle the data and scientific workflow as well?
Containers for Reproducible Research
Reproducible research is a vast and active conversation (Gentleman and Lang 2007; Peng 2011; LeVeque, Mitchell, and Stodden 07/2012; Stodden, Leisch, and Peng 2014; Stodden and Miguez 2014; Meng et al. 07/2015; Marwick 2016; Meng and Thain 2015) far out of scope for this paper. Although containers are a boon for managing software, currently the most active and vibrant conversation around software containers for science (and other academic disciplines) revolves around containers’ potential application for reproducible research. Encapsulation enable researchers to capture the full fidelity of their research environments and portability allows sharing beyond prosaic methods sections in journal articles. Containers seem like a natural fit for reproducible research; the capability of containers to encapsulate the software dependencies of a computational research environment can be extended to include the data, metadata, code, and workflow of a specific project or publication. In such a case, a container transforms from a universal workbench into a product or publication oriented object with all of the dependencies and content in a single bundle.
The conversation around containers for reproducibility is very active with informal workshops bringing together a variety of disciplines.[39] In reviewing the literature, no single discipline can be singled out as leading the adoption of containers for reproducible research, this work crosses disciplinary lines. Researchers from multiple disciplines, brought together by methodological commonalities (specifically a DevOps approach) to achieve reproducibility. Certain kinds of computational researcher, especially those drawing on industrial data science, incorporate ideas from the software development industry, such as continuous integration[40] to create reproducible computational workflows (Beaulieu-Jones and Greene 2016). These approaches take specific technologies like Docker and combine them with cutting edge software development techniques to automate reproducibility. Again, the distinguishing feature of researchers talking about and using containers are those with the DevOps approach to their work, not any individual discipline as a whole.
One early contribution that kicked off the conversation about containers and reproducibility is Karl Boettiger’s (2015) article, An Introduction to Docker for reproducible research. Boettiger enumerates several technological barriers to reproducible research:
- Dependency hell, which we discussed above and is a major motivating factor for the adoption of containers in the high performance research computing community.
- Imprecise documentation, highlights the fact that documentation is poor, especially all of the technical and procedural details that are left out of a publication’s method section
- Code rot, the recognition that software and its dependencies are not static, but dynamic entities continually being updated with bug fixes, new features, and security patches. Such changes can sometimes impact the reproducibility of computational research.
- Lack of adoption, there are already many existing technical solutions for creating reproducible research, but they are narrow or heavy handed solutions.
Boettiger argues Docker provides a solution to these technical problems by encapsulating dependencies, explicit documentation in Dockerfiles, avoiding code rot through versioned container images and enabling adoption by being portable, lightweight, and easy to integrate into existing workflows. However, he recognizes technical remedies alone will not solve the problems of reproducible research. There are significant cultural barriers to overcome, not in the least convincing researchers to be more transparent and share their code and workflow. Boettiger points out the incentive structures do not exist to reward the additional work of sharing additional materials like code and data. Docker or other container technologies add another burden if researchers are not already accustomed to a DevOps approach to their practice.
Both a benefit and a challenge for containers is the lack of standardized ways to express workflows within containers. While some argue the Dockerfile provides explicit documentation of a container’s contents, the execution of processes inside a container lack explicit semantics. Software containers are somewhat agnostic to the semantics expression of their contents, which potentially makes each individual container a black box.[41] This makes a container is a blank canvas within which researchers can pour dependencies, code, data, and metadata, which has an important advantage because many people have their own personal workflows and environments (especially in the absence of collaborators). This “blank canvas” approach may be a boon for early adoption as computational research practices change, but a thousand and one bespoke containers for a thousand and one research projects still presents problems for reproduction and preservation. There have been efforts to standardize the expression of workflows within the container (O’Connor et al. 2017) and Knoth (2016) uses Docker encapsulate a standardized workflow building on a standard software install for the genomics community.
How data and code get into containers and how to execute a workflow is unique for each container because the technology is so endlessly flexible. While this affords easier adoption because it imposes fewer constraints upon how the computational work gets done, it may present challenges to the long-term reproducibility of the encapsulated research.
Containers for Preservation
The question of preserving containers is a natural follow-on from containers for reproducible research. This conversation has only just begun with only some very preliminary work specifically focused on the preservation of containers as opposed to more general conversations about scientific workflows. There has been some initial effort to propose a conceptual framework for the preservation of Docker containers (Emsley and De Roure 2017), leveraging linked-open-data standards to provide a semantic expression of the workflow. Such efforts are a good start, but significantly more work is needed. The DASPOS project,[42] an NSF funded effort to address the problems of data and software preservation for science, convened a workshop to explicitly discuss containers for software preservation.
Containers are seen, alongside virtual machines, as one mechanism for preserving scientific software (Thain, Ivie, and Meng 2015). Scientific software preservation is related to container preservation and projects like SoftwareX and ReproZip are fellow travelers of the preservation landscape.[43] Other efforts like Collective Knowledge and the Occam use containers, but focus their attention on preserving the components and build process with richer semantics, so the encapsulated workflow could be rebuilt on whatever the next technology may emerge after containers.[44]
Part and parcel to preservation is standardization. For long-term preservation to succeed, some formal and agreed upon standards must be established. In industry, standardization of containers is being driven by the Open Container Initiative (OCI). The OCI specifications for runtime environments and image formats just recently reached 1.0.[45] Unfortunately, this is entirely an industry driven effort and it is unclear if the unique needs of academic applications are being addressed. Singularity, the container developed by and for academia/HPC has initiated their own effort towards a format specification, in part to address the lack of best practices for content (data files, code, and metadata) placed inside containers.[46]
The harsh reality, which any archivist or digital preservation professional would be quick to point out, is thinking about preservation is never at the forefront of researcher’s or system administrator’s attention. Archivists and librarians are often dealing with unmanaged dumps of data and information in a variety of formats that haven’t been designed with preservation in mind. This will continue to be true with software containers. Researchers are already using a variety of container technologies (Docker, Shifter, Singularity, etc.) and there will probably never be universal agreement on a standard format or set of practices. Just like the way in which archives get email or document dumps, we can anticipate they may someday get container image dumps. “Without information on the environment, source code and other relevant metadata, we can inspect it but don’t have a can opener.” - (Mooney and Gerrard 2017). Digital forensics tools like BitCurator[47] give archivists such a can opener, but they don’t yet support software containers.
There are still many open questions related to the preservation of containers:
- How to deal with proprietary, licensed software?
- How to deal with proprietary hardware like GPUs?
- What additional environmental information is needed?
- For layered container images are the lower layers available?
- Does it make more sense to focus preservation efforts on the components rather than the container?
- How do we preserve Docker or other runtime engines?[48]
There are not enough archivists and digital preservation professionals participating in the academic conversations around software containers. Researchers need to invite archivists to help with the preservation of their work, but also the archives community needs to start actively paying attention to the rapidly moving technological landscape around them; this is a mutual failure. Computer scientists are taking up the task of digital preservation without consulting on the wealth of expertise from the archives community. While this problem is far out of scope for this document, it is relevant because software containers solve some preservation problems, but also introduce new ones. Furthermore, the challenges of digital preservation and reproducible research are not exclusively technological and computer scientists are not necessarily equipped to deal with many of the social, historical, and political dynamics of preservation.
Scientists, scholars, and researchers are under enormous pressure publish their research, this is how they get credit and this is how the incentive structure of academia operates. The additional work of making data sharable, documenting code, and producing reproducible workflows is unrewarded overhead work. This problem is even worse for digital scholarship and digital publications that do not have a traditional print-centric publication as the expression of the work. All of this work, the workflows or non-traditional publications do not enter the systems of publishing and so they often not given due credit and are certainly not preserved. But what if they were? What if we thought about containers as publications?
Full-Stack Scholarship
This section of the report considers containers as publications, which is a radical idea whose promises and perils have not been fully evaluated. This section is speculative and the ideas are very much “in beta.” The potential for containers to be publications in and of themselves is precisely what we want to open up for further discussion.
There is some existing work considering the possibility of using containers as a basis for scholarly publishing. Opening Reproducible Research,[49] is a promising model being developed by geoscientists at the University of Münster. The project has been developing a conceptual model, a platform, and standards for executable research compendia (ERC), which are publishable units that include “the actual paper, source code, the computational environment, the data set, and a definition of a user interface.”[50]
ERC leverage the affordances of containers and lay out a set of standardized requirements for their contents. In this model, the container is just one part of a collection of files that get wrapped into a standard archival format familiar to librarians.[51] The scope of ERC are very modest, they are meant for small research workflows that can be run on a laptop using programming languages like R. Workflows requiring big data or multi-mode high performance computing are not their target audience.
Beyond what goes inside the container, the project proposes a publication and review process that is compatible with traditional scholarly publishing. ERC are meant to integrate into traditional journal publishing processes because they presuppose a print-centric paper as the main output. They are meant as a way to augment traditional publications by wrapping up all of the additional materials related to the production of a traditional, print-centric publication and necessary for reproducibility.[52]
ERC is an interesting model for thinking about containers are publications and their implementation could provide a basis for further work. However, the final publication in their workflow is still static, print-centric documents. Furthermore, the execution of an ERC is expected to time bound (even if it takes a long time) and have clearly defined outputs. This is at odds with some of the use cases for digital projects in the humanities that don't have such clearly delineated boundaries between data, workflow, and publication and are long-running processes. For example, a database backed web application such as Infinite Ulysses is a long running process and doesn't necessarily fit the ERC model describe above.[53] ERC is not, in its current design, supportive of multi-modal humanities (McPherson 2009).
There are other efforts thinking about the rich expressions. Brett Victor’s Media for Thinking the Unthinkable[54] has inspired a new genre of multimodal publications that leverage the affordances of the web browser.[55] One of the complications of web publishing is the difficulties with encapsulation and portability, websites can have porous boundaries and they belie the very idea of portability, they exist at one location and are difficult to move because of the conflation of address and identifier.[56] There is the potential for a fruitful marriage between the affordances of web and the affordances of containers to create executable, media rich documents that are encapsulate and portable.
In “Computational Publishing with Jupyter” Andrew Odewahn combines Jupyter Notebooks with software containers as a model for computational publishing.[57]
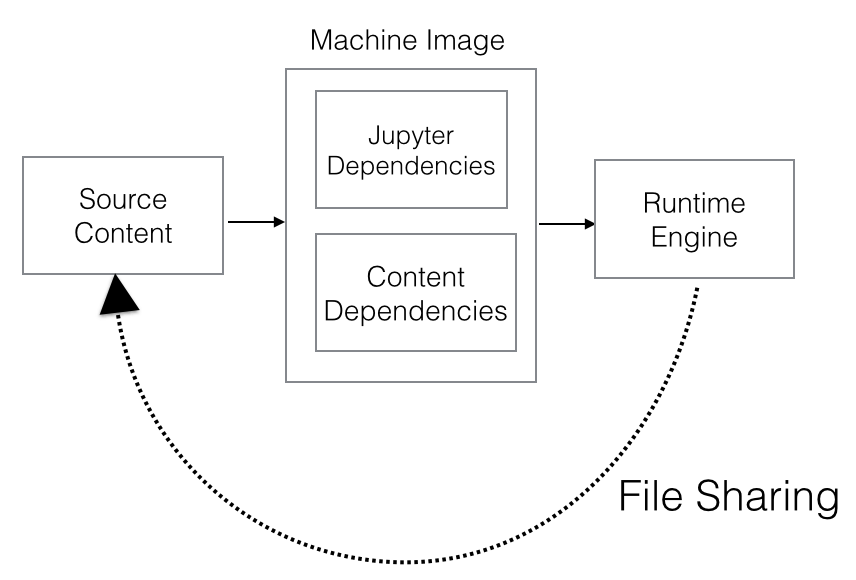
Odewahn’s model outlines the basic components for thinking about software containers as publications that encapsulate Jupyter Notebooks as a web-based interface for accessing and interacting with the container’s contents. We can generalize this model and think more abstractly about maintaining a distinction between content and platform, yet still encapsulating both.
Drawing on this model we can think of computational publications as having the following components:
- Source Content - This is the meat of a computational publication and also the most recognizable. Source content could be narrative, data, code and/or other digital components.
- A Container Image - Container images, would encapsulate the source content, but also two sets of dependencies:
- Platform dependencies - Computational publications will need a platform in which to provide an interface to the content. Odewahn uses Jupyter as the platform, but we could expand to consider a full range of expressive platforms, for example Bookworm, Omeka, Scalar, static sites, or custom web applications.[59] These platforms each have their own set of software dependencies.
- Content dependencies - Source content may include a separate set of dependencies from those of the expressive platform. A researcher’s Jupyter Notebook may require visualization libraries like matplotlib, altair, or plotly and machine learning libraries like scikit-learn. These are not requirements of the Jupyter platform, but are necessary for executing the source content.
- The Container Engine - The previous components related to the runtime environment internal to the computational publication, the container engine is the external dependency and environment for running the publication. The container engine can be designed to execute any computable publication conforming to a standardized interface.
This rough, high-level model is provided to stimulate discussion and thinking around the technical architecture of computational publications. It may already be evident that conceptualizing digital scholarship in this way would require many in academia to shift in both thinking and practice. Although many digitally savvy practitioners in academic contexts already partition and document their software with the expectation of future use, switching from a non-containerized approach to containerization would require a shift in practice comparable to moving from a no-collaboration-expected model to an applications that assumes third-party developers.
Yet, if a scholar were to “rethink the entire enterprise of academic publishing” and rebuild “public scholarship from the ground up ,” it would behoove that scholar to examine the things we take for granted, such as publishers, peer-review, preservation, and the support structures that have evolved around the affordances of print materiality (Trettien 2017). Such a shift will have much greater ramifications than simply our technological choices, it changes the role and relationship with the many invisible laborers of scholarly publishing like librarians and archivists. The potential benefits of such reconceptualization would be manifold, but advocating such a shift raises obvious complications about power, academic prestige, and the material labor of who maintains new publishing infrastructures.
Conclusion: Containers for Digital Scholarship
Software containers’ adoption in academic research contexts leverage three technological affordances:
- Encapsulation - Containers encapsulate everything an application needs to execute in a single logical unit, the image.
- Portability - With standard image formats and runtime interfaces, containers can be easily moved and executed across different platforms and infrastructures.
- Performance - Because containers share resources with the host operating system they are faster and lighter than alternatives like virtual machines.
The most popular container technology, Docker, has become the de facto standard for packaging and deploying enterprise applications like web servers and databases in the commercial sector. While Docker is also popular amongst researchers, academically developed alternatives like Singularity have emerged to address some the the problems unique to the research context, such as security..
- How are containers being discussed and adopted in academic research contexts?
In scanning the literature, public discourse, and conference/workshop discussions the adoption and use of software containers roughly falls into three use cases. First, containers are being used by system administrators, especially in high performance computing, to reduce the complexity of scientific software dependencies. Second, beyond encapsulating software dependencies, containers can bundle data and workflows to enable reproducible research. Third, the encapsulation of software, data, and workflows means they are not only portable across space, but also time making containers an important format for the long-term preservation of research workflows and outputs.
- Which aspects of containerization have not yet been fully explored in the context of digital scholarship?
The current discussion and application of containers has primarily focused on alleviating existing problems in the maintenance, reproduction, and preservation of computational research. We argue containers open a new horizon of possible practices in scholarly publishing. While there are some efforts thinking about such possibilities, the conceptual models and the use-cases are still conservative. Publishing environments and workflows of a traditional print-centric publication does not go far enough, we think containers could be an enabling format for a new breed of computational publications.
The value proposition of containers in academia is not merely that they run fast; encapsulate information; or are portable from a laptop to a supercomputer. The technological problems containers solve are a result of social and organizational differences across academia.[60] Solomon Hykes, the CEO of Docker, has also made the point that “the real value of Docker is not technology” but rather “getting people to agree on something.”[61] Regardless of whether academic community settles upon Docker or Singularity as the container technology of choice, the more important point is to have standards that (mostly) everyone can agree upon. The emergence and adoption of standards are complicated social, technological, cultural, political, and economic processes with a complicated tangle of agendas, incentives, and artifacts (Egyedi 2001; Millerand and Bowker 2007; Lampland and Star 2009; Busch 2011). There is a lot of social and conceptual work, beyond the technical work of building platforms, as various disciplines and constituencies adopt or resist the potential changes that software containers make possible in scholarly publishing.
Understanding the risks and rewards of software containers is an important next step for this project. The second stage of “Digits: a Platform to Facilitate the Production of Digital Scholarship,” will involve creating a second report for publication. This follow-up document will focus on assessing the infrastructural needs of digital humanists around publishing and preserving web-centric digital scholarship and will evaluate the potential of software containers for specific disciplinary practices in the humanities.
Bibliography
- Beaulieu-Jones, Brett K., and Casey S. Greene. 2016. “Reproducible Computational Workflows with Continuous Analysis.” bioRxiv, August, 056473.
- Belmann, Peter, Johannes Dröge, Andreas Bremges, Alice C. McHardy, Alexander Sczyrba, and Michael D. Barton. 2015. “Bioboxes: Standardised Containers for Interchangeable Bioinformatics Software.” GigaScience 4: 47.
- Boettiger, Carl. 2015. “An Introduction to Docker for Reproducible Research.” ACM SIGOPS Operating Systems Review 49 (1): 71–79.
- Busch, Lawrence. 2011. Standards: Recipes for Reality. MIT Press.
- Chung, M. T., N. Quang-Hung, M. T. Nguyen, and N. Thoai. 2016. “Using Docker in High Performance Computing Applications.” In 2016 IEEE Sixth International Conference on Communications and Electronics (ICCE), 52–57.
- Clark, Dav, Aaron Culich, Brian Hamlin, and Ryan Lovett. 2014. “BCE: Berkeley’s Common Scientific Compute Environment for Research and Education.” In Proceedings of the 13th Python in Science Conference (SciPy 2014).
https://www.researchgate.net/profile/Dav_Clark/publication/290855636_BCE_Berkeley’s_
Common_Scientific_Compute_Environment_for_Research_and_Education/links/
569c560c08aeeea985a5b390.pdf. - “CoreOS.” 2017. Accessed July 27. https://coreos.com/rkt.
- “Deltron 3030 - Virus Lyrics | MetroLyrics.” 2017. Accessed July 30. http://www.metrolyrics.com/virus-lyrics-deltron-3030.html.
- Devisetty, Upendra Kumar, Kathleen Kennedy, Paul Sarando, Nirav Merchant, and Eric Lyons. 2016. “Bringing Your Tools to CyVerse Discovery Environment Using Docker.” F1000Research 5 (December): 1442.
- “Docker.” 2017. Docker. Accessed July 27. https://www.docker.com/.
- “Docker Alternatives and Competitors | G2 Crowd.” 2017. G2 Crowd. Accessed July 27. https://www.g2crowd.com/products/docker/competitors/alternatives.
- Egyedi, Tineke. 2001. “Infrastructure Flexibility Created by Standardized Gateways: The Cases of XML and the ISO Container.” Knowledge, Technology & Policy 14 (3): 41–54.
- Emsley, I., and D. De Roure. 2017. “A Framework for the Preservation of a Docker Container.” In . https://ora.ox.ac.uk/objects/uuid:f567f27a-4efb-431e-abcb-07b6e8c03ce2.
- Felter, W., A. Ferreira, R. Rajamony, and J. Rubio. 2015. “An Updated Performance Comparison of Virtual Machines and Linux Containers.” In 2015 IEEE International Symposium on Performance Analysis of Systems and Software (ISPASS), 171–72.
- Gentleman, Robert, and Duncan Temple Lang. 2007. “Statistical Analyses and Reproducible Research.” Journal of Computational and Graphical Statistics: A Joint Publication of American Statistical Association, Institute of Mathematical Statistics, Interface Foundation of North America 16 (1): 1–23.
- Hale, Jack S., Lizao Li, Chris N. Richardson, and Garth N. Wells. 2016. “Containers for Portable, Productive and Performant Scientific Computing.” arXiv:1608.07573 [cs], August. http://arxiv.org/abs/1608.07573.
- Holdgraf, Chris, Aaron Culich, Ariel Rokem, Fatma Deniz, Maryana Alegro, and Dani Ushizima. 2017. “Portable Learning Environments for Hands-On Computational Instruction: Using Container- and Cloud-Based Technology to Teach Data Science.” In Proceedings of the Practice and Experience in Advanced Research Computing 2017 on Sustainability, Success and Impact, 32. ACM.
- Hosny, Abdelrahman, Paola Vera-Licona, Reinhard Laubenbacher, and Thibauld Favre. 2016. “AlgoRun: A Docker-Based Packaging System for Platform-Agnostic Implemented Algorithms.” Bioinformatics 32 (15): 2396–98.
- Hung, Ling-Hong, Daniel Kristiyanto, Sung Bong Lee, and Ka Yee Yeung. 2016. “GUIdock: Using Docker Containers with a Common Graphics User Interface to Address the Reproducibility of Research.” PloS One 11 (4): e0152686.
- Jacobsen, D. M., and R. S. Canon. 2016. “Shifter: Containers for HPC.” In Cray Users Group Conference (CUG’16).
- Jacobsen, Douglas M., and Richard Shane Canon. 2015. “Contain This, Unleashing Docker for Hpc.” Proceedings of the Cray User Group. http://ai2-s2-pdfs.s3.amazonaws.com/77d9/7e17c7129a810d14fb8dfd17fa4ca07e18bc.pdf.
- Kamvar, Zhian N., Margarita M. López-Uribe, Simone Coughlan, Niklaus J. Grünwald, Hilmar Lapp, and Stéphanie Manel. 01/2017. “Developing Educational Resources for Population Genetics in R: An Open and Collaborative Approach.” Molecular Ecology Resources 17 (1): 120–28.
- Knoth, C., and D. Nust. 2016. “Enabling Reproducible OBIA with Open-Source Software in Docker Containers.” In http://proceedings.utwente.nl/456/.
- Lampland, Martha, and Susan Leigh Star. 2009. Standards and Their Stories: How Quantifying, Classifying, and Formalizing Practices Shape Everyday Life. Cornell University Press.
- Le, Emily, and David Paz. 2017. “Performance Analysis of Applications Using Singularity Container on SDSC Comet.” In Proceedings of the Practice and Experience in Advanced Research Computing 2017 on Sustainability, Success and Impact, 66. ACM.
- LeVeque, Randall J., Ian M. Mitchell, and Victoria Stodden. 07/2012. “Reproducible Research for Scientific Computing: Tools and Strategies for Changing the Culture.” Computing in Science & Engineering 14 (4): 13–17.
- Levinson, Marc. 2008. The Box: How the Shipping Container Made the World Smaller and the World Economy Bigger. Princeton University Press.
- Marwick, Ben. 2016. “Computational Reproducibility in Archaeological Research: Basic Principles and a Case Study of Their Implementation.” Journal of Archaeological Method and Theory, January, 1–27.
- McPherson, T. 2009. “Introduction: Media Studies and the Digital Humanities.” Cinema Journal 48 (2): 119–23.
- Meng, Haiyan, Rupa Kommineni, Quan Pham, Robert Gardner, Tanu Malik, and Douglas Thain. 07/2015. “An Invariant Framework for Conducting Reproducible Computational Science.” Journal of Computational Science 9: 137–42.
- Meng, Haiyan, and Douglas Thain. 2015. “Umbrella: A Portable Environment Creator for Reproducible Computing on Clusters, Clouds, and Grids.” In Proceedings of the 8th International Workshop on Virtualization Technologies in Distributed Computing, 23–30. VTDC ’15. New York, NY, USA: ACM.
- Millerand, F., and G. C. Bowker. 2007. “Metadata Standards: Trajectories and Enactment in the Life of an Ontology’.” Formalizing Practices: Reckoning with Standards, Numbers and Models in Science and Everyday Life.
- Mooney, James, and David Gerrard. 2017. “Software ‘Best Before’ Dates: Posing Questions about Containers and Digital Preservation.” presented at the Docker Containers for Reproducible Research Workshop, Cambridge, June 28. https://drive.google.com/file/d/0B7Jaz2j9AIcWTlRSakNxY2hNdVE/view.
- Moreews, François, Olivier Sallou, Hervé Ménager, Yvan Le bras, Cyril Monjeaud, Christophe Blanchet, and Olivier Collin. 2015. “BioShaDock: A Community Driven Bioinformatics Shared Docker-Based Tools Registry.” F1000Research, December. doi:10.12688/f1000research.7536.1.
- Nüst, Daniel, Markus Konkol, Edzer Pebesma, Christian Kray, Marc Schutzeichel, Holger Przibytzin, and Jörg Lorenz. 2017. “Opening the Publication Process with Executable Research Compendia.” D-Lib Magazine 23 (1/2). doi:10.1045/january2017-nuest.
- O’Connor, Brian D., Denis Yuen, Vincent Chung, Andrew G. Duncan, Xiang Kun Liu, Janice Patricia, Benedict Paten, Lincoln Stein, and Vincent Ferretti. 2017. “The Dockstore: Enabling Modular, Community-Focused Sharing of Docker-Based Genomics Tools and Workflows.” F1000Research 6 (January): 52.
- Peng, R. D. 2011. “Reproducible Research in Computational Science.” Science 334 (6060): 1226–27.
- Portworx, 2019 Container Adoption Survey, Presented by Portworx and Aqua Security. https://portworx.com/wp-content/uploads/2019/05/2019-container-adoption-survey.pdf
- Portworx, 2019 Container Adoption Survey, Presented by Portworx and Aqua Security. https://portworx.com/wp-content/uploads/2018/12/Portworx-Container-Adoption-Survey-Report-2018.pdf
- Špaček, František, Radomír Sohlich, and Tomáš Dulík. 2015. “Docker as Platform for Assignments Evaluation.” Procedia Engineering 100: 1665–71.
- Stodden, Victoria, Friedrich Leisch, and Roger D. Peng. 2014. Implementing Reproducible Research. CRC Press.
- Stodden, Victoria, and Sheila Miguez. 2014. “Best Practices for Computational Science: Software Infrastructure and Environments for Reproducible and Extensible Research.” Journal of Open Research Software 2 (1): 8.
- Szitenberg, Amir, Max John, Mark L. Blaxter, and David H. Lunt. 2015. “ReproPhylo: An Environment for Reproducible Phylogenomics.” PLoS Computational Biology 11 (9): e1004447.
- Thain, Douglas, Peter Ivie, and Haiyan Meng. 2015. “Techniques for Preserving Scientific Software Executions: Preserve the Mess or Encourage Cleanliness?” doi:10.7274/R0CZ353M.
- Trettien, Whitney. 2017. “A Feminist Note on ‘Publication, Power, and Patronage.’” Medium. Medium. July 27. https://medium.com/@whitneytrettien/a-feminist-note-on-publication-power-and-patronage-a834ed6a5cd0.
- Verma, Abhishek, Luis Pedrosa, Madhukar Korupolu, David Oppenheimer, Eric Tune, and John Wilkes. 2015. “Large-Scale Cluster Management at Google with Borg.” In Proceedings of the Tenth European Conference on Computer Systems, 18:1–18:17. EuroSys ’15. New York, NY, USA: ACM.
- “What Is DevOps? - Amazon Web Services (AWS).” 2017. Amazon Web Services, Inc. Accessed July 31. https://aws.amazon.com/devops/what-is-devops/.
- Williams, Jason J., and Tracy K. Teal. 01/2017. “A Vision for Collaborative Training Infrastructure for Bioinformatics: Training Infrastructure for Bioinformatics.” Annals of the New York Academy of Sciences 1387 (1): 54–60.
- Willis, Craig, Mike Lambert, Kenton McHenry, and Christine Kirkpatrick. 2017. “Container-Based Analysis Environments for Low-Barrier Access to Research Data.” In Proceedings of the Practice and Experience in Advanced Research Computing 2017 on Sustainability, Success and Impact, 58. ACM.