A PCR Assay Detects Legionella pneumophila Harboring Mobile Element ICE–βox in a Variety of Water Sources
Skip other details (including permanent urls, DOI, citation information)
: This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 United States License. Please contact mpub-help@umich.edu to use this work in a way not covered by the license.
For more information, read Michigan Publishing's access and usage policy.
Introduction
The most common waterborne disease in the United States is the pneumonia Legionnaire’s disease,[1] with a case fatality rate as high as 80%.[3] Legionella pneumophila is readily detected in the built environment,[2] and epidemiologic surveys suggest that 63% to 84% of US hospital water systems are colonized with Legionellae.[4] In a study of 209 Paris hospital water systems, chlorination treatment correlated with increased prevalence of Legionellae.[5] In summer 2015, contaminated cooling towers in Bronx, New York, sickened 128 and killed 12. Even after disinfection, 15 of 35 towers still tested positive for this pathogen.13 Thus L. pneumophila persistence in the built environment is a public health concern.
L. pneumophila acquires traits via horizontal gene transfer. Integrative conjugative elements (ICEs) are mobile genetic elements that are either integrated into bacterial chromosomes or excised as episomes.[6] Excision requires direct repeat nucleotide sequences called attachment (att) sites. Recipient bacteria must harbor the att site to maintain a newly acquired ICE (Figure 1). The att sites align during recombination and facilitate mobility. This process leaves 1 att site on the chromosome and 1 on the ICE episome, allowing for reintegration after excision events. Thus those bacteria that do not carry an att site cannot integrate an incoming ICE into the bacterial genome.
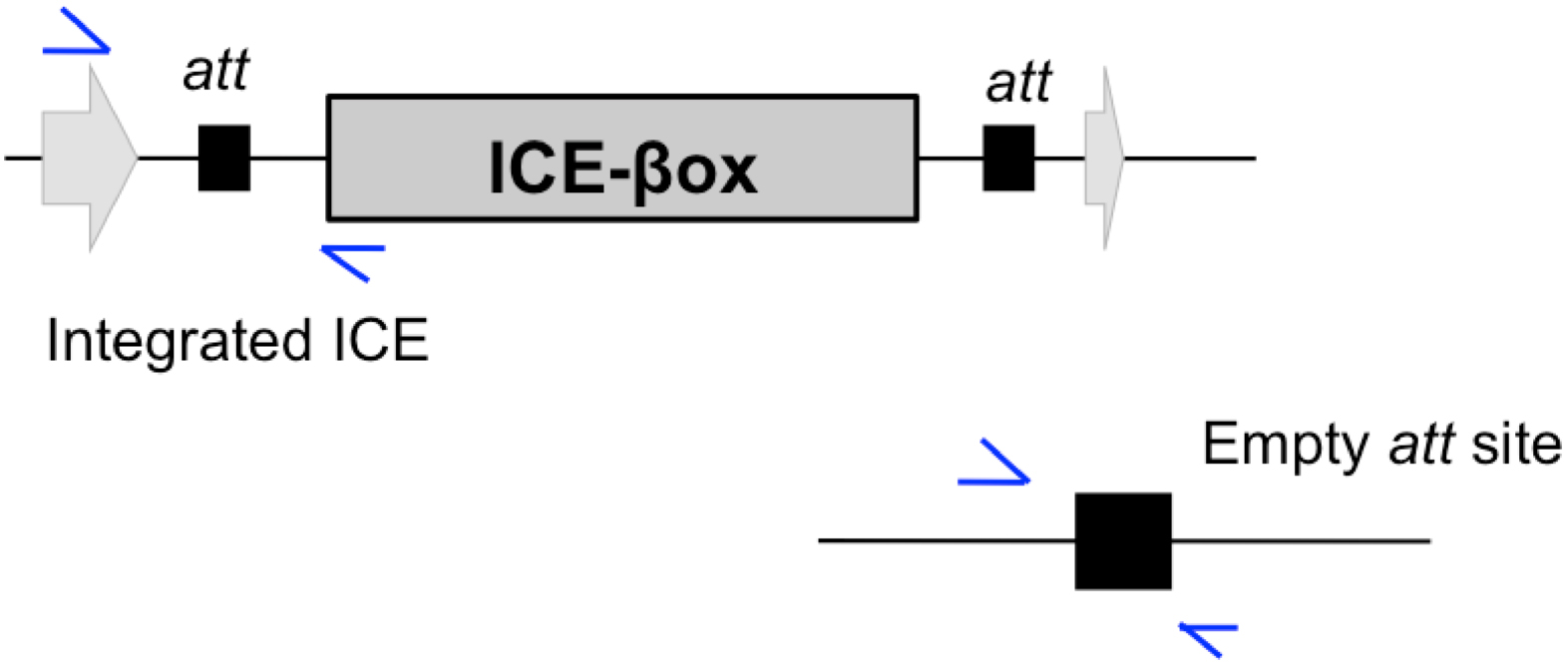
ICE–βox of L. pneumophila strain Philadelphia–1 confers resistance to ß-lactam antibiotics, oxidative stress encountered within macrophages, and bleach.[7] As chlorine disinfectants are the primary method of eradicating waterborne pathogens, the discovery of a mobile element with these fitness advantages is concerning. Given the prevalence of L. pneumophila isolated from chlorinated environments, it is possible these disinfectants select for resistant isolates harboring ICE–βox. In this way, water treatment may actually select for more fit strains of the pathogen it is designed to eradicate.[8],[9]
To probe the relationship between disinfection and oxidative stress resistance, we designed a multiplex polymerase chain reaction (PCR) screen to assess ICE–βox presence in clinical and environmental L. pneumophila isolates. This screen is specifically designed to detect either the presence of integrated ICE–βox or the att site necessary to acquire the element in 1 reaction. In this study, we aimed to determine the prevalence of ICE–βox in L. pneumophila clinical and environmental isolates using this assay.
Methods
Bacterial strains
One hundred eighty–three L. pneumophila isolates representing serogroups 1 through 17 were selected from the United States Centers for Disease Control and Prevention (CDC) Legionella reference collection to determine the prevalence of ICE–βox and the att site. Included were typing strains representing several serogroups, clinical isolates, built environment isolates linked to particular infection cases (Table 2, column 4), and 1 natural water isolate with no known exposure to disinfectants. The positive control strain for this assay was L. pneumophila strain Philadelphia–1 derivative Lp02, known to contain ICE–βox, and the negative control strain was the derivative JR32, known to lack the element.[7]
DNA extraction
Genomic DNA was isolated at the CDC using the InviMag Bacterial DNA kit/KFmL (Invitek, Hayward, California) on the KingFisher mL (Thermo Fisher Scientific, Philadelphia, Pennsylvania), MagNA Pure Compact (Roche, Basel, Switzerland), or EZ1 advanced XL (Qiagen, Hilden, Germany) platforms according to manufacturer’s guidelines.
Real–Time Multiplex PCR
Real–Time Multiplex PCR was performed on 1 ng/ul DNA using Quanta PerfeCTa Multiplex qPCR SuperMix (Quanta Biosciences, Gaithersburg, Maryland) on the ABI7500 Standard platform (Applied Biosciences). Specific primers were designed manually to amplify the integrated form of ICE–βox, the empty att site, or a pan–genome control as described previously[7] (Table 1, Figure 1). Primers were verified using genomic DNA isolated from L. pneumophila strain Lp02 as a positive control and strain JR32 as a negative control.[7]
| Primer Name | Sequence (5′ → 3′) |
|---|---|
| Philadelphia-F | CGGAATAGACCAGACCCAAATGGCGCG |
| Paris-F | AGCCGGAATAGACCGATTAAAAATG |
| Lens-F | TTGGGGAAGAGCCTTTTAAATGG |
| Lorraine-F | AATAATGTGGGGGTTTACTAAATGGC |
| HL-F | ATGCAAATTAAATCAACAAAGTGGC |
| Alcoy-F | AATTGGGAAAGAGCCATTAAATGGC |
| Sg12-F | GATTTTAAAAGGATTAAATGGCG |
| Integrated-R | GATTTGATGCATCGTAAGTTGTTGATT |
| Empty-R | ATAAAATGTTCATCCACACCCCAT |
| Integrated-ABY-P | ABY-TGTTTTCTATTATTGAGTATCAG-MGBNFQ |
| Empty-TX-RED-P | TX615-CGCTCGTAGCTCAGCTGGATAGAGTACTT-BHQ2 |
| Pan-Leg-F | GGCGACCTGGCT TC |
| Pan-Leg-R1 | GGTCATCGTTTGCATTTATATTTA |
| Pan-LegFAM P | FAM-ACGTGGGTTGCAA-MGBNFQ |
Results
To pilot our surveillance strategy, we screened 183 clinical and built environment L. pneumophila strains isolated from outbreaks sent to the CDC using primers specifically designed to amplify the integrated ICE–βox or the empty att site. This screening assay proved to be sequence–specific, as the PCR products were of the expected size and did not detect ICE–βox or the att site in negative control strains. Of the 183 isolates, 57 (31.1%) contained integrated ICE–βox, and the remaining 126 (68.9%) carried its att site (Table 2). One hundred thirteen were serogroup 1 strains, the most common serogroup associated with infection. Of these, 24/84 (28.5%) of clinical isolates and 22/29 (75.8%) of built environment strains carried ICE–βox, and the remainder contained att (Table 2).
Although knowledge of the water treatment protocols used in the environments represented is limited, of the 3 outbreak locations where data exist, chlorine disinfectant concentrations ranged from 0.2 to 0.7 ppm Cl2 (Table 2). In the 1 natural water isolate tested in this initial study, att, but not ICE–βox, was detected (Table 2). Compared to serogroup 1 strains, ICE–βox was less prevalent for both clinical (8.2%) and environmental (29.4%) nonserogroup 1 strains.
| Isolate Source | Contains ICE-βox | Contains att Site Only | Location Examples |
|---|---|---|---|
| Clinical | 24/84 (28.6%) | 60/84 (71.4%) | Lung, sputum, bronchial wash |
| Built environment | 22/29 (75.8%) | 7/29 (24.1%) | Cooling tower, faucet, fountain |
| Cl2 exposure | 2/3 (66.6%) | 1/3 (33.3%) | Source treated with 0.2 to 0.7 ppm Cl2 |
| Natural environment | 0/1 (0%) | 1/1 (100%) | Soil and outdoor shower in Thailand |
| Non-Sg1 clinical | 4/49 (8.2%) | 45/49 (91.8%) | Lung, sputum, bronchial wash |
| Non-Sg1 environmental | 5/17 (29.4%) | 12/17 (70.6%) | Showerhewad, tap water |
| Total | 57/183 (31.1%) | 126/183 (68.9%) |
Conclusion
This pilot study demonstrates that a multiplex PCR assay can detect both integrated ICE–βox and its att site in a range of L. pneumophila serotypes and isolates. Since all 183 strains tested contained the ICE–βox att site, it appears that the capacity to accept and stably integrate ICE–βox is widespread in L. pneumophila, regardless of serotype.
ICE–βox was more prevalent in built environment samples than in clinical isolates (75.8% vs 28.5%, respectively, P < 0.001). The apparent increased frequency of ICE–βox in environmental isolates is consistent with the hypothesis that disinfectant–treated water selects for strains that carry ICE–βox. The 0.2 to 0.7 ppm Cl2 exposure levels reported for 3 of the environmental isolates are much lower than the 2 ppm chlorine level ICE–βox confers protection to.[7] To continue to assess whether water treatment impacts ICE–βox prevalence, we are next keen to compare ICE–βox prevalence in larger sets of isolates subjected to known disinfectant treatments with natural water L. pneumophila isolates with no known exposure to chlorinated chemicals. With this multiplex PCR assay in hand, we are poised to determine the potential for the spread of disinfectant– and antibiotic–resistant L. pneumophila in the built environment.
Acknowledgments
This work was supported by the Endowment for Basic Sciences at the University of Michigan Medical School (MSS), a University of Michigan Rackham Merit Fellowship (KJF), and the Molecular Mechanisms of Microbial Pathogenesis training program (NIH T32 AI007528; KJF).
References
Brunkard JM, Ailes E, Roberts VA, et al. Surveillance for waterborne disease outbreaks associated with drinking water—United States, 2007–2008. MMWR Surveill Summ. September 23 2011;60(12):38–68.

van Heijnsbergen E, Schalk JA, Euser SM, et al. Confirmed and potential sources of Legionella reviewed. Environ Sci Technol. April 1 2015.

Phin N, Parry–Ford F, Harrison T, et al. Epidemiology and clinical management of Legionnaires’ disease. Lancet Infect Dis. 2014;14(10):1011–1021.

Yu PY, Lin YE, Lin WR, et al. The high prevalence of Legionella pneumophila contamination in hospital potable water systems in Taiwan: implications for hospital infection control in Asia. Int J Infect Dis. July 2008;12(4):416–420.

Merault N, Rusniok C, Jarraud S, et al. Specific real–time PCR for simultaneous detection and identification of Legionella pneumophila serogroup 1 in water and clinical samples. Appl Environ Microbiol. March 2011;77(5):1708–1717.

Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. August 2010;8(8):552–563.

Flynn KJ, Swanson MS. Integrative conjugative element ICE–βox confers oxidative stress resistance to Legionella pneumophila in vitro and in macrophages. mBio. 2014;5(3):e01091–01014.





Allen HK, Donato J, Wang HH, et al. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol. April 2010;8(4):251–259.

Segura PA, Francois M, Gagnon C, Sauve S. Review of the occurrence of anti–infectives in contaminated wastewaters and natural and drinking waters. Environ Health Perspect. May 2009;117(5):675–684.


